Designed for developers
The MZKit project its source code is open sourced on github and totally free of charge for used in your academic project.
Mzkit is an open source raw data file toolkit for mass spectrometry data analysis, provides by the BioNovoGene corporation. The features of mzkit inlcudes: raw data file content viewer(XIC/TIC/Mass spectrum plot/MS-Imaging), build molecule network, formula de-novo search, de-novo annotation of the unknown metabolite features, MALDI single cell metabolomics data analysis, pathological slide viewer and targeted data quantification.
The MZKit project its source code is open sourced on github and totally free of charge for used in your academic project.
MZKit is easily to learn and also work in fast speed. You also can running the MZKit R# script in parallel for deal with the large scale metabolomics raw data files.
The development of the MZKit workbench software is mainly address of the metabolomics raw data processing and raw data visualization.
The MZKit workbench has a user friendly UI, which means you don't needs to do some scripting programming for run data analysis of your mass spectrum raw data file.
You can create a mass spectrum data analysis pipeline on a windows server via scripting in R# programming language. The MZKit software is written in VisualBasic.NET language, and you also can build a software by using the MZKit library in your .NET program.
Watch the MZKit tutorial video on BILIBILI which we created for the audience like you.
MZKit Video Tutorials from “六神无主鸠”
There ares some awesome features of MZKit workbench that wait for discovered by you. Download and
install MZKit and then try to find out them!


MS-Imaging data visualization of your biological samples in MZKit workbench can use for compares of your multiple targeted metabolite compounds on one graphics canvas. Such data visualization of the metabolite can visual and compares of the spatial enrichment results for the multiple targeted drugs or metabolism products for your experiment design. It is useful for presenting the biomarker in a spatially way by MS-imaging.
Visit Data Visualization Gallery to learn more.
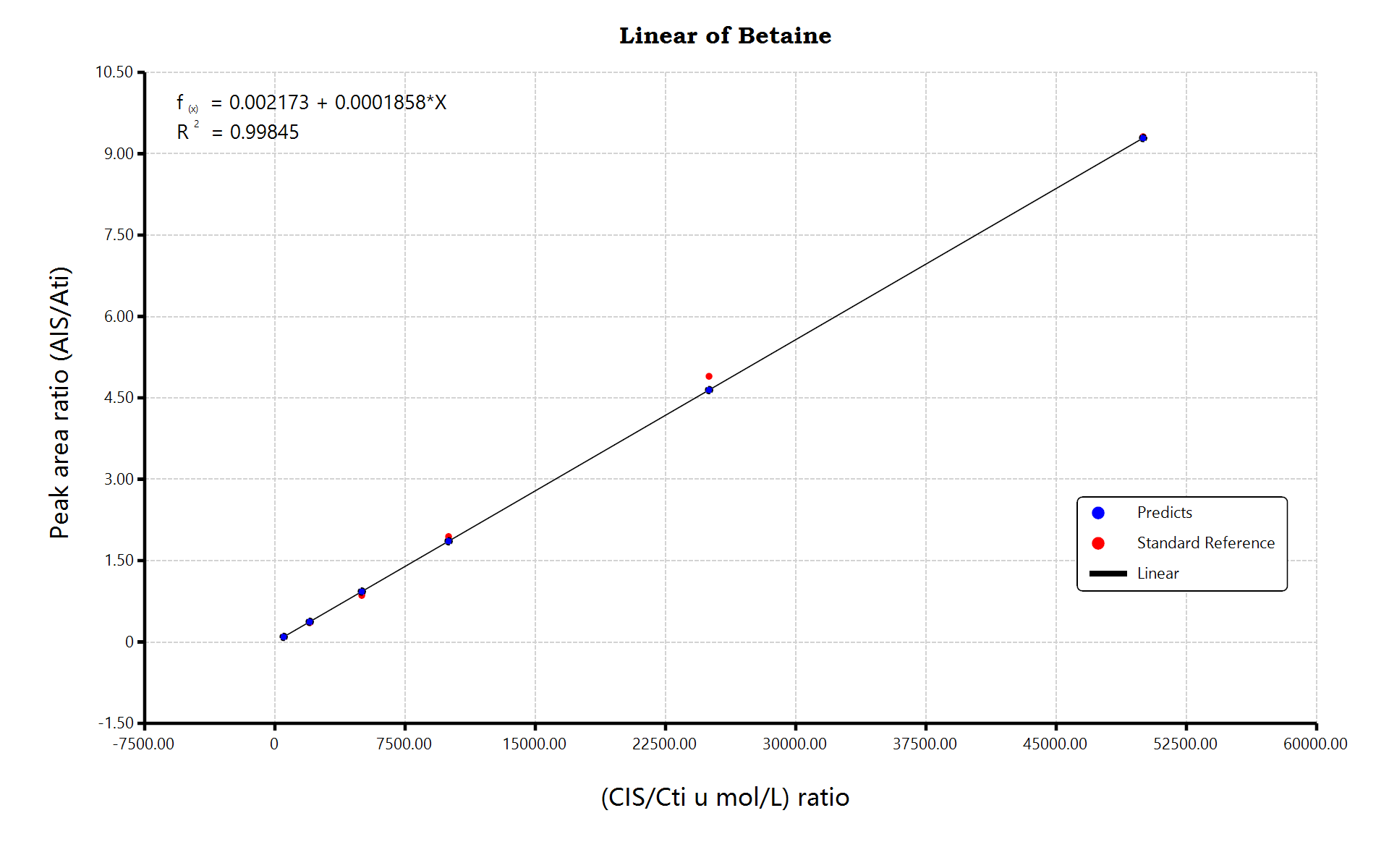
Targeted metabolomics is the study and analysis of specific metabolites, and it take parts in disease research, animal model verification, biomarker discovery, disease diagnosis, drug development, drug screening, drug evaluation, clinical research and plant metabolism research:
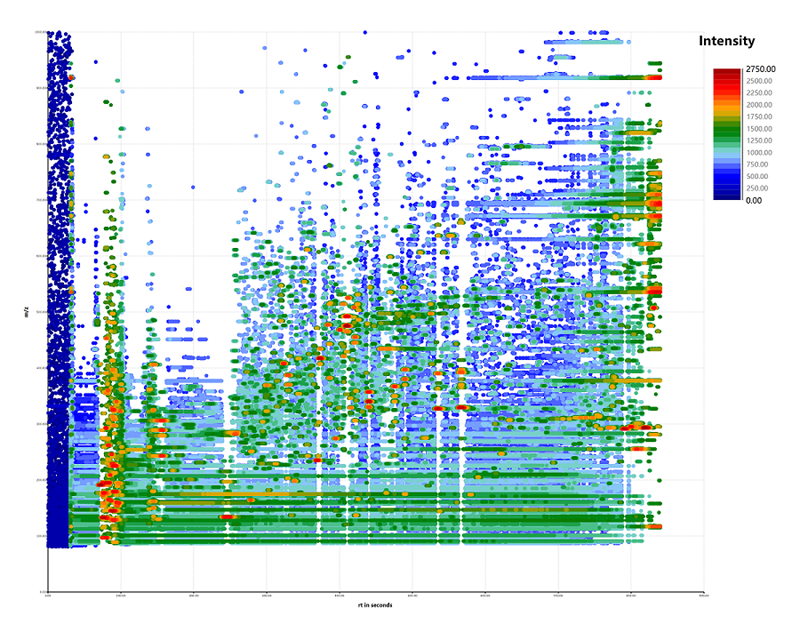
Untargeted metabolomics is a "discovery mode" process and it relies on differential comparison between groups of samples (for example cases versus controls): Non-targeted metabolomics can analyze metabolites derived from the organisms comprehensively and systematically. It is an unbiased metabolomics analysis that can discover new biomarkers.
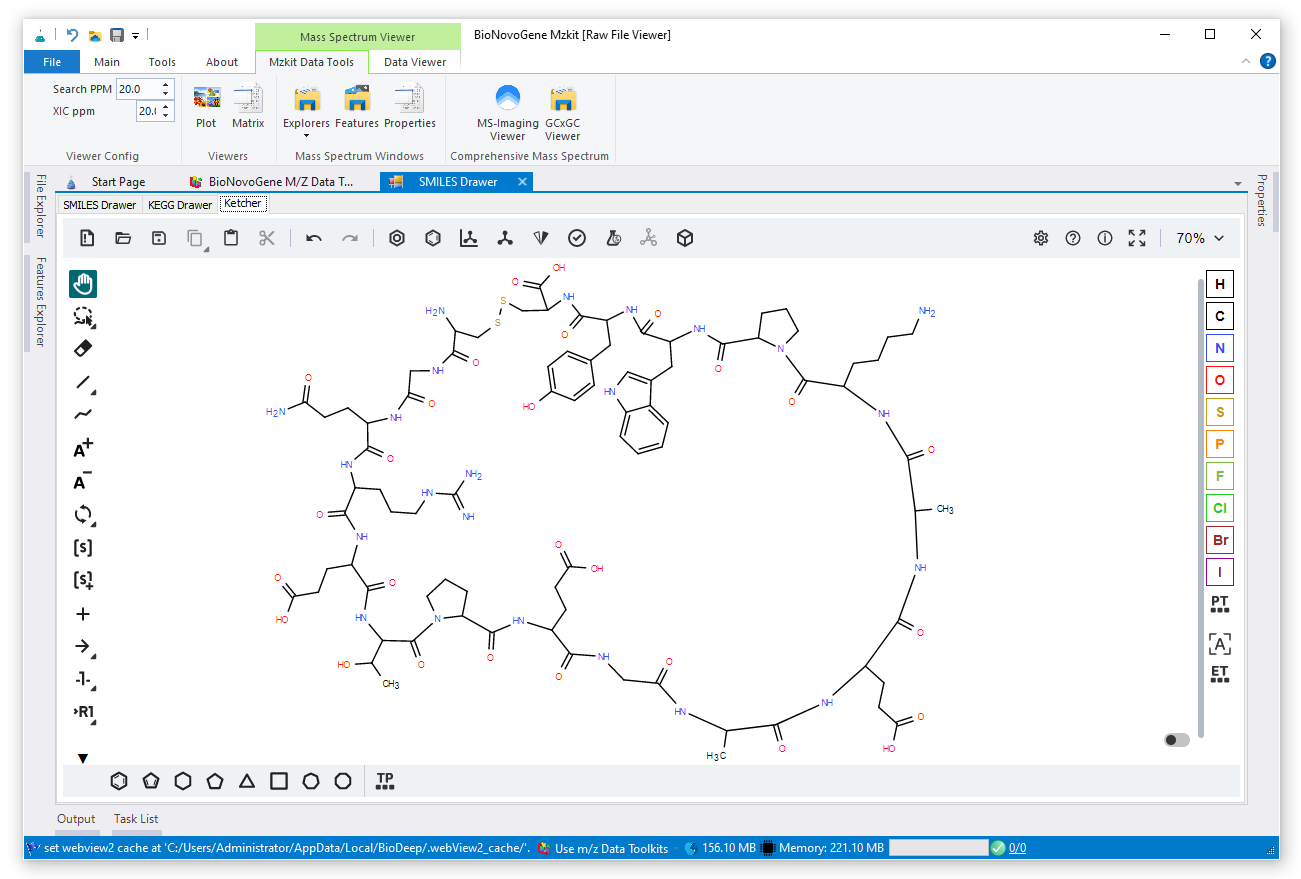
Draw chemical molecule structure via Ketcher in MZKit, and then convert the molecule structure data as the SMILES/InChI string. Then we could do mass spectrum prediction in MZKit via calling CFM-ID tool for generates the in-silicon MS2 matrix, finally search your MS2 matrix inside multiple sample that which has been loaded into the MZKit workspace.
MZKit workbench software required of .NET Framework 4.8 runtime:
https://dotnet.microsoft.com/download/dotnet-framework/net48
Rstudio application for run mzkit R# automation pipeline required of .NET Core 6.0(windows-x64)
runtime:
https://dotnet.microsoft.com/en-us/download/dotnet/6.0
The Microsoft WebView2 runtime(windows-x64) is required for some interactive data visualization and
analysis report generation:
https://developer.microsoft.com/en-us/microsoft-edge/webview2/
Unidata netCDF-C library is required for read GCxGC or GCMS netCDF rawdata file:
https://downloads.unidata.ucar.edu/netcdf-c/4.9.2/netCDF4.9.2-NC4-64.exe
MZKit workbench supports Windows 10/11 64Bit system and Windows Server 2016/2019/2022.
Hardware Requirement:
You can cite MZKit in your literature work if needed: xie, guigang &
BioNovoGene. BioNovoGene
mzkit: Data toolkits for processing NMR, MALDI MSI, LC-MS and GC-MS raw data, chemoinformatics data
analysis and data visualization. (2022) doi:10.5281/zenodo.7040586.
Copyright (c) 2025 xieguigang@metabolomics.ac.cn / gg.xie@bionovogene.com, BioNovoGene Co., LTD.
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the "Software"), to deal
in the Software without restriction, including without limitation the rights
to use, copy, modify, merge, publish, distribute, sublicense, and/or sell
copies of the Software, and to permit persons to whom the Software is
furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all
copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR
IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY,
FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER
LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM,
OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE
SOFTWARE.
The development of the MZKit workbench library based on these works, you are also can cite these works if you use MZkit software: